|
Leffingwell &
Associates
|
Alchemist WebPick Awarded by the webzine of ChemWeb.com  |
|
General Physiology of Olfaction Trigeminal Sense in the Olfactory Epithelium The Odorant Binding Proteins Odorant receptors The Cellular Membrane G-Protein Coupled Receptors G-Proteins The cAMP Transduction Cascade Ion Protein Channels Other Second Messengers in Olfaction - cGMP, IP3, NO, CO Chemical Olfactory Stimulation - Theories on Olfaction The Steric Theory of Odor The Vibrational Theory of Odor Vibrational Induced Electron Tunneling Spectroscope Theory Ribonucleotides as the Odorant carrier? Recent Events in Olfactory Understanding A Combinatorial Process for odor Interpretation Combinatorial Process Visualization Human Olfactory Receptor Genes Enantiomeric Specificity in the Olfactory bulb |
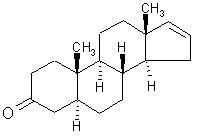
Olfaction-Page 2 John C. Leffingwell, Ph.D. The Odorant Binding Proteins In 1979, Steven Price and coworkers5 discovered a protein in the olfactory epithelium that bound the chemical "anisole, while Fesenko, et.al.6, found a camphor binding protein. Since that time a number of other so-called "Odorant Binding Proteins" or "OBP’s" have been found for odorant chemicals such as benzaldehyde (cherry-almond odor), 2-Isobutyl-3-methoxypyrazine (green bell pepper odor) and 5-a-androst-16-en-3-one (urine odor).  .............................. ..............................
5-a-ANDROST-16-EN-3-ONE Although the role of Odorant Binding Proteins is not totally clear, one proposed role is that they bind lipophilically to odorants in the aqueous/lipid mucous increasing the concentration and then facilitate transport through the mucous layer to the receptors in the olfactory membrane. Other possibilities suggested are that they bind to the ligand and receptor and assist in transport across the olfactory membrane, or that they act as a kind of filter to prevent excessive amounts (over saturation) from reaching the receptor. The role for odorant binding proteins became clearer in 2000 with the characterization of human lipocalins involved in odorant binding. Lipocalins are carrier proteins for hydrophobic molecules in many biological fluids. In the oral sphere (nasal mucus, saliva, tears), they have an environmental biosensor function and are considered involved in the detection of odours and pheromones. Human OBP (IIa) was strongly expressed in the nasal structures, salivary and lachrymal glands, and lung, therefore having an oral sphere profile while hOBP (IIb) was more strongly expressed in genital sphere organs such as the prostate and mammary glands.6a Similarly, although monoclonal antibodies (MCAB) had been shown to bind odorants (anisole & benzaldehyde) by Price in 19886b, in 1999 workers at the Pluckthun lab6c (Univ. Zurich) showed monoclonal antibodies were elicited against the small hydrophobic hapten traseolide (a commercially available musk, 5-acetyl-1,1,2,6-tetramethyl-3-isopropylindane (Traseolide) . Antibody variable region sequences were found to belong to different sequence groups, and the binding characteristics of the corresponding antibody fragments were studied. To elucidate the structural basis for the fine specificity of binding, they determined the crystal structure of the fragment complexed with the Traseolide at 2.6 A resolution. The crystal structure showed that only van der Waals interactions are involved in binding. The structural understanding of odorant specificity in an antibody gives a degree of insight in the physical principles on how specificity for such hydrophobic molecules may be achieved.
Odorant receptors. In 1991, Linda Buck7,9 and Richard Axel8,9 discovered both the family of transmembrane proteins that were believed to be the odor receptors and some of the genes that encode them. They cloned and characterized 18 different members of an extremely large multigene family that encodes the seven transmembrane proteins whose expression was restricted to the olfactory epithelium. This was a seminal breakthrough in our potential understanding of the olfactory system9 which led to them receiving the Nobel Prize in Physiology in 2004.9a The proteins found all contained the 7 helical transmembrane structure and contained sequence similarity to other members of the "G-protein" linked receptor family. [See below for details on membranes and G-protein coupled receptors]. Lancet's group in Israel and Zozulya and coworkers at Senomyx have shown there are about 350 odorant receptor genes and about 560 odorant receptor pseudogenes in humans.7, 12f, 12c, 12g,12h This number of genes and pseudogenes, specific to the olfactory system, comprises nearly 2% of the 50,000 or so genes of the human genome. This number is second only to the receptors of the immune system.
Until early 1998, however, there was no direct proof that functionally these were actually odor receptors. In January, 1998, Firestein10 and coworkers at Columbia and Yale University effectively demonstrated that genes coded to produce olfactory receptors could be inserted into the rat olfactory system and that specific odorant chemicals would generate significantly higher signaling as measured by the electrical activity in the neurons.11 "The Columbia team inserted two linked genes, one that codes for a rat olfactory receptor, called rat I7, and a gene for green fluorescent protein (GFP), a substance found normally in fluorescent jellyfish but now used by molecular biologists to mark genetically altered cells, into a disabled adenovirus - the same virus that causes colds. The modified adenovirus was in turn introduced into rat olfactory neurons. Cells that carried the rat I7 gene also carried the GFP gene, and could be discerned because they glowed bright green when exposed to blue light. Firestein's graduate student, Haiqing Zhao, now at Johns Hopkins Medical School, treated rats with the modified adenovirus and then exposed their olfactory neurons to various odorants. He monitored the electrical activity in the neurons, producing a chart called an electro-olfactogram. Electrical activity was highest when the nerve cells were exposed to octanal, an aldehyde that smells meaty to humans".12 {Note that most flavorists & perfumers would not describe octanal as being "meaty", but as being "fatty-fruity with citrus-orange notes"] In this work researchers evaluated 74 odorants on a specific odor receptor. A long standing question as to whether individual receptors recognize multiple odorants, or do single neurons have multiple receptors now appears clarified.12a It appears that to reconcile the ability of organisms to detect far more than 1,000 discrete odors, the odors must participate in some kind of combinatorial processing: i.e., one receptor must be able to interact with several discrete odorants. Conversely, an odor molecule must be capable of interacting with multiple receptors. By inference, an individual odor will activate multiple glomeruli in the olfactory bulb.12b This is discussed in more detail at the end of this review. By 2000, Doron Lancet & co-workers at the Weizmann Institute of Science Crown Human Genome Center had constructed a database of human olfactory receptor genes by a highly automated data mining system12c, 12h to detect all OR genes present in the public databases. This non-redundant dataset includes 855 human OR genes, of which 391 have full open reading frame. For details and searchable OR gene data, click on the picture below.  The authors of HORDE (The Human Olfactory Receptor Data Exploratorium), where this originates from, indicate that olfactory receptor genes are present in practically all human chromosomes, with only chromosomes 20 and Y being somewhat devoid of ORs. Chromosome 11 is by far the richest in OR genes. This type data also may help elucidate the human evolutionary tree.12d, 12i In June, 2001, Zozulya, Echeverri & Nguyen of Senomyx published a paper entitled "The human olfactory receptor repertoire" in which they reported the identification and physical cloning of 347 putative human full-length odorant receptor genes. Comparative sequence analysis of the predicted gene products allowed them to identify and define a number of consensus sequence motifs and structural features of this vast family of receptors. They believe that these sequences represent the essentially complete repertoire of functional human odorant receptors.12f .They found some differences between their data and those in the HORDE database, however. These include 29 "human OR" (hOR) genes that were apparently identified as pseudogenes in the HORDE database, but encoded as functional hOR candidates in their analysis, as well as 10 hORs not found at that time in HORDE. This online article includes a downloadable file with all 347 hOR's in FASTA format.12g In the January 22, 2002 issue of Nature Neuroscience12j, Stuart Firestein with Xinmin Zhang at Columbia University identified the mouse OR genes from the nearly complete Celera mouse genome by a comprehensive data mining strategy. They found 1,296 mouse OR genes (including 20% pseudogenes). Human ORs cover a similar 'receptor space' as the mouse ORs, suggesting that the human olfactory system has retained the ability to recognize a broad spectrum of chemicals even though humans have lost nearly two-thirds of the OR genes as compared to mice. With elucidation of the general classification of olfactory receptors as being G-protein coupled receptors, let us now examine how such receptors work (in general) and whether such receptors can be influenced. In order to set the stage, however, we must first examine the structure of the cellular membranes in which the receptors reside. The Cellular Membrane Below is depicted a biological membrane transversed by an olfactory receptor protein. The membrane consists of a phosholipid bilayer. At the both the extracellular and intracellular sides depicted in green are the hydrophilic negatively charged phosphate groups. The "tails" of the phospholipids consist of hydrocarbon chains (fat) which orient towards each other (pointing inward to the middle of the membrane), creating a hydrophobic environment. These hydrocarbon "tails" are depicted in gray. Such membrane lipid bilayer are virtually impermeable to large molecules and relatively impermeable to charged ionic molecules and yet quite permeable to lipid soluble low molecular weight molecules. 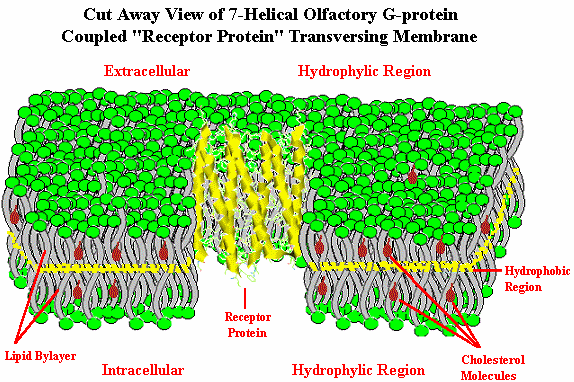
The "Fluid Mosaic Model" of the biological membrane accounts for the fact that the membrane includes proteins and cholesterol, as well as phospholipids and other molecules, and is fluid…not static. The membrane is about 5 nanometers thick, yet individual phospholipids diffuse rapidly throughout the two dimensional surface of the membrane. It is known that phospholipids can move to the opposite side of the membrane within a few minutes at room temperature in bacterial cells even though the distance is several thousand times the size of the phospholipid. Proteins also diffuse (or move fluidly) within the membrane, but at a much slower rate due to their massive size. The early work leading to this model has been described by S. Jonathan Singer.12k Cholesterol is a necessary component of these cellular membranes helping to break up Van der Waals interactions and the close packing of the lipid portion of the phospholipids, which is integral to making the membrane more fluid. Membranes serve to (1.) define and compartmentalize the cell, (2.) serve as the locus of specific functions, (3.) control movement of substances into and out of the cell and its compartments and (4.) play a pivotal role in cell-to-cell communication and detection of external signals (e.g., olfaction). _______________ 5. Goldberg S., J. Turpin and S. Price, Anisole binding protein from olfactory epithelium: evidence for a role in transduction, Chem. Senses & Flavour, 4:207 (1979). 6. Fesenko E.E., V.I. Nonoselov and L.D. Krapivinskaya, Molecular mechanisms of olfactory reception, Biochim. Biophys. Acta, 587:424 (1979). 6a. Lacazette E, Gachon AM, Pitiot G., A novel human odorant-binding protein gene family resulting from genomic duplicons at 9q34: differential expression in the oral and genital spheres, Hum Mol Genet, Jan 22;9(2):289-301 (2000) 6b. Price, S., Willey, A., Effects of antibodies against odorant binding proteins on electrophysical responses to odorants, Biochem Biophys Acta, 965: 127ff (1988) 6c. Langedijk AC, Spinelli S, Anguille C, Hermans P, Nederlof J, Butenandt J, Honegger A, Cambillau C, Pluckthun A., Insight into odorant perception: the crystal structure and binding characteristics of antibody fragments directed against the musk odorant traseolide, J Mol Biol 1999 Oct 1;292(4):855-69 6d. Tegoni, Mariella, Roberto Ramoni, Enrico Bignetti, Silvia Spinelli, and Christian Cambillau. "Domain swapping creates a third putative combining site in bovine odorant binding protein dimer." Nature Structural & Molecular Biology 3, no. 10 (1996): 863-867. 6e. Pelosi, Paolo, Rosa Mastrogiacomo, Immacolata Iovinella, Elena Tuccori, and Krishna C. Persaud. "Structure and biotechnological applications of odorant-binding proteins." Applied microbiology and biotechnology 98, no. 1 (2014): 61-70. 6f. Jeong, Yong Taek, Jaewon Shim, So Ra Oh, Hong In Yoon, Chul Hoon Kim, Seok Jun Moon, and Craig Montell. "An odorant-binding protein required for suppression of sweet taste by bitter chemicals." Neuron 79, no. 4 (2013): 725-737. 7. Linda Buck: Information coding in the vertebrate olfactory system, Annual review of neuroscience. 1996 Mar;19(1):517-44; Linda Buck: Unraveling the sense of smell (Nobel lecture)." Angewandte Chemie International Edition 44 (38) (2005): 6128-6140 8. Richard Axel: "The molecular logic of smell." Scientific American 273, no. 4 (1995): 154-159., Richard Axel: Scents and sensibility: a molecular logic of olfactory perception (Nobel lecture). Angewandte Chemie International Edition, 44(38) (2005): 6110-6127. 9. Buck, L. and R. Axel, A novel multigene family may encode odor recognition: a molecular basis for odor recognition, Cell, 65:175 (1991). 9a. http://www.nobelprize.org/nobel_prizes/medicine/laureates/2004/ 10. http://www.columbia.edu/cu/biology/faculty/firestein/index.html 11. Zhao, H., L.Ivic, J.M. Otaki, M. Hashimoto, K. Mikoshiba, S. Firestein, Functional expression of a mammalian odorant receptor. Science, Jan 9;279(5348):237-242 (1998) 12. Columbia University Record, 23(12), Jan.23, 1998; http://www.columbia.edu/cu/record/23/12/16.html 12a. Travis, John. Making Sense of Scents, Science News, April 10, 1999, Malnic B, Hirono J, Sato T, Buck LB; Combinatorial receptor codes for odors. Cell, Mar 5;96(5):713-23 (1999): http://www.hhmi.org/news/buck.htm 12b. Ronhi, A. Maureen, Chemical & Engineering News, Dec. 23, 1996 12c. Glusman G., Bahar A., Sharon D., Pilpel Y., White J, Lancet D., The olfactory receptor gene superfamily: data mining, classification, and nomenclature., Mamm. Genome, Nov;11(11):1016-23 (2000). 12d. Lancet, Doron, et.al., Molecular recognition and evolution in biological repertoires: from olfaction to the origin of life (2000) at http://www.weizmann.ac.il/Biology/open_day_2000/images/lancet.pdf 12f. Sergey Zozulya, Fernando Echeverri & Trieu Nguyen, The human olfactory receptor repertoire, http://genomebiology.biomedcentral.com/articles/10.1186/gb-2001-2-6-research0018 12g. Zozulya, et. al., http://genomebiology.biomedcentral.com/articles/10.1186/gb-2001-2-6-research0018 12h. Glusman G, Yanai I, Rubin I, Lancet D., The complete human olfactory subgenome, Genome Res. 2001 May;11(5):685-702 12i. Fuchs T, Glusman G, Horn-Saban S, Lancet D, Pilpel Y, The human olfactory subgenome: from sequence to structure and evolution, Hum Genet 2001 Jan;108(1):1-13; See also Sharon, D., Glusman, G., Pilpel, Y., Horn-Sabarn, S. and Lancet, D., 1998. Genome Dynamics, Evolution, and Protein Modeling in the Olfactory Receptor Gene Superfamily, Annals of the New York Academy of Sciences, 855(1), pp.182-193. 12j. Zhang X, Firestein S., The olfactory receptor gene superfamily of the mouse, Nat Neurosci., Feb;5(2):124-133 (2002); See also Godfrey, Paul A., Bettina Malnic, and Linda B. Buck. "The mouse olfactory receptor gene family." Proceedings of the National Academy of Sciences 101, no. 7 (2004): 2156-2161. 12k. Singer, S. J., Some Early History of Membrane Molecular Biology, Annual Review of Physiology. Vol. 66: 1-27, 2003; http://www.annualreviews.org/doi/pdf/10.1146/annurev.physiol.66.032902.131835 ____________
|
|
|
Telephone: 01-770-8895111 - Email: leffingwell@mindspring.com
TERMS OF SERVICE.............PRIVACY POLICY